

No-code AI training tool for 3D medical images (DICOM RT/CT/MR) is newly released as an additional feature of Growth RTV. Medical professionals can easily develop and apply AI using their own data without programming.
(*In the initial release, we will provide a function for developing AI for automatic extraction of 3D regions, which is the most in demand, and will support medical AI for other applications in due course.)
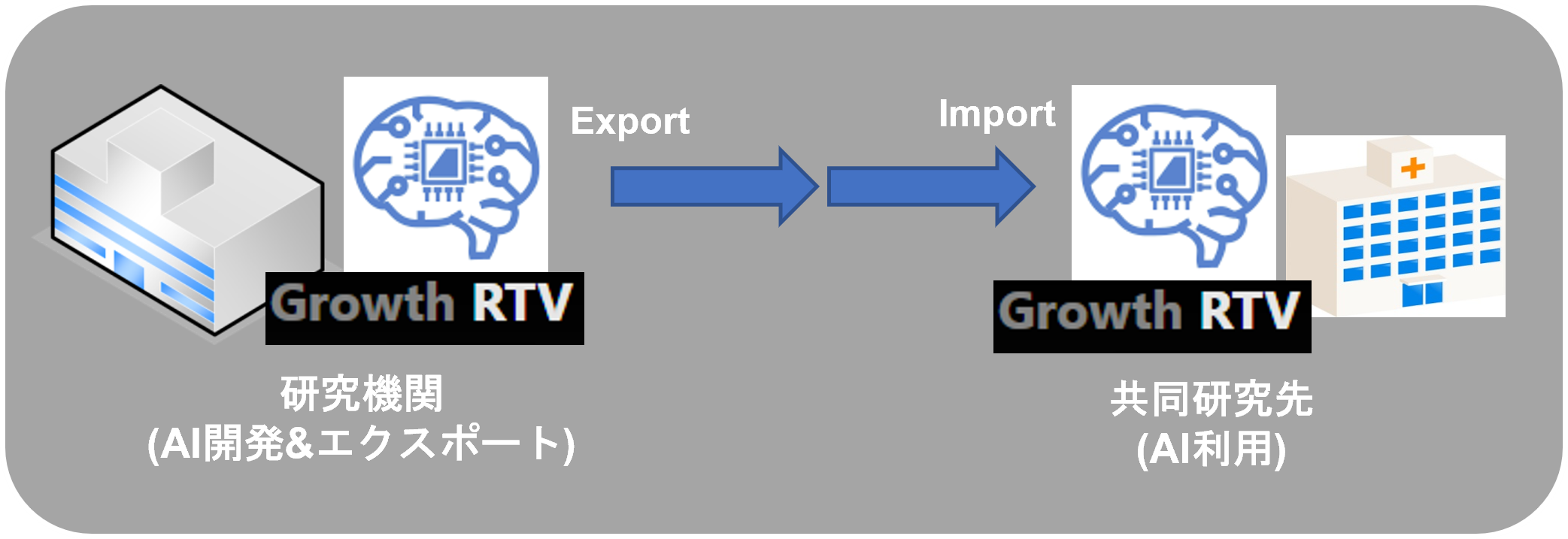
The trained AI and execution commands can be passed between users, and by using the export/import function of Growth RTV, they can be provided to “Growth RTV” users in other departments or facilities for use in a wide range of use cases.
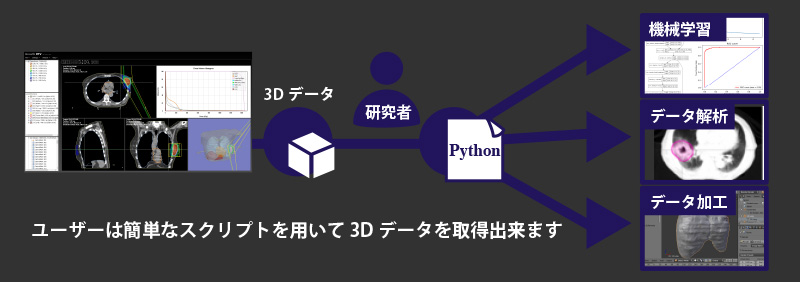
The 3D medical image deep learning support platform with programmable Python integration functions allows users to acquire 3D image data with simple Python scripts and perform data annotation, data analysis, data extraction and processing, learning, etc. for deep learning without knowing how to handle DICOM. data annotation, data analysis, data extraction, processing, learning, etc. for deep learning. The platform also provides a seamless environment for the development and use of medical AI, allowing users to add new organ regions to CT and DICOM-RT data, and to reflect the AI developed by the research team in DICOM data for subjective evaluation.
For more information
We will begin trial sales of our self-developed “AI-based CT metal artifact reduction technology” as a cloud service for non-medical applications such as modeling, education, and research.
For more information
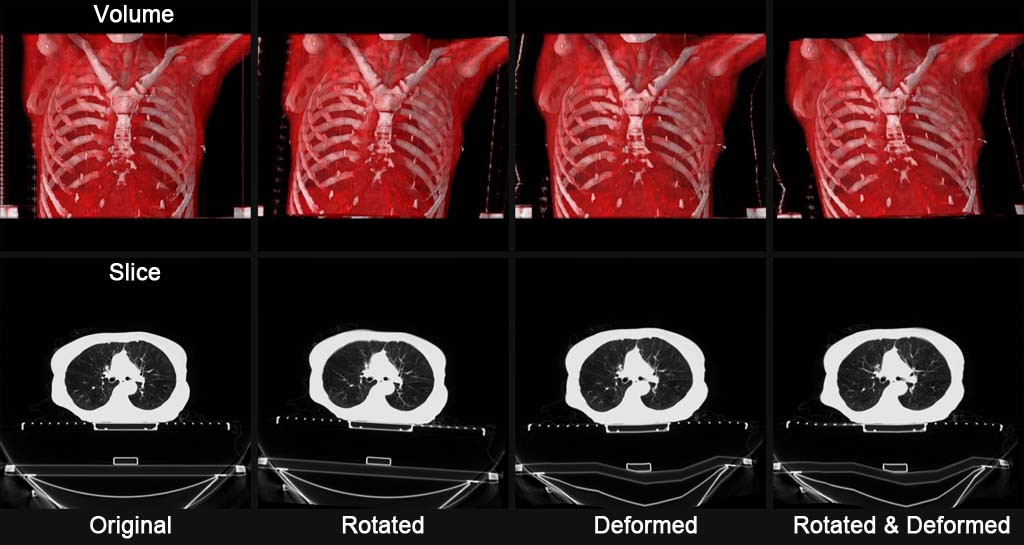
The Python platform is equipped with a data expansion function for 3D volume images by deformation and rotation. Users can easily call this function using the Python language to expand data such as CT, MRI, and organ regions for deep learning. The internal processing engine of this function implements high-speed completion processing using the CUDA language, enabling continuous generation of a large amount of extended data in a short time.
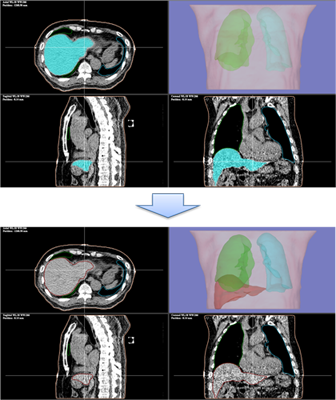
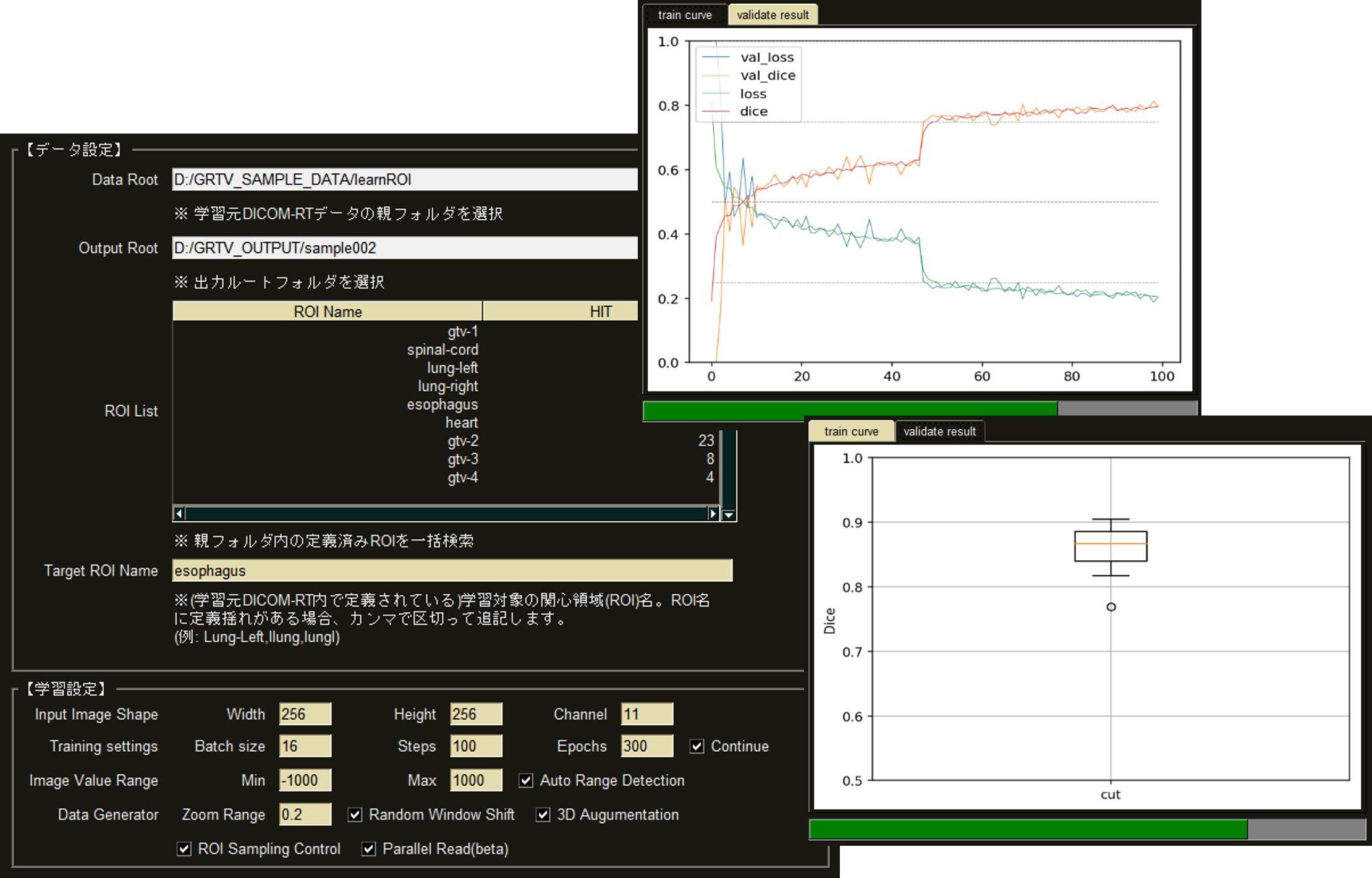
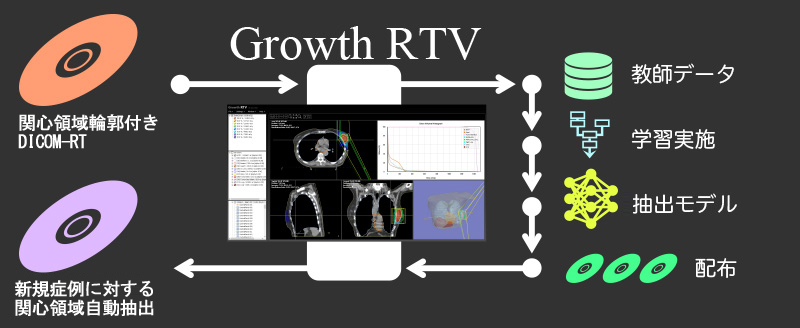
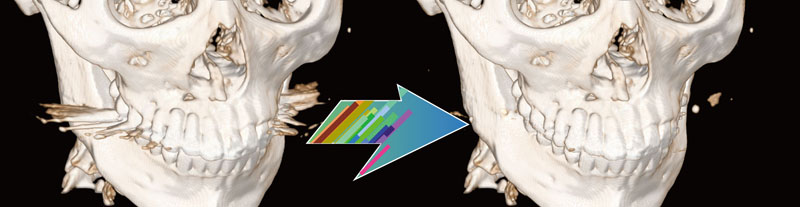
By extracting organ regions using our originally developed DICOM-RT analysis framework and using it for deep learning, 3D regions of target organs can be extracted with high accuracy in a single click.
Currently, extraction models for thorax and abdominal organs (body surface, lungs (L/R), liver, kidneys (L/R), and spinal canal) have been installed, and other parts will be added through successive updates.
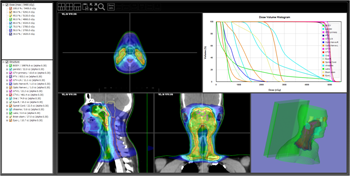
It supports not only CT and MRI radiological image DICOM, but also DICOM-RT format, which is radiological image data, and is equipped with substantial basic display functions such as each section display, 2D and 3D contour display, irradiation field display, and dose distribution display.
List of display functions:
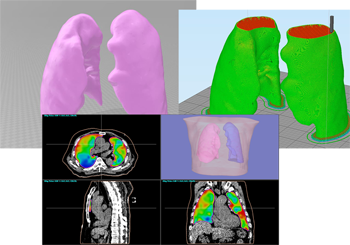
The extracted organ shapes can be exported in STL, PLY, and DICOM-RT Structure Set formats, and can be linked with 3D printers for stereolithographic modeling and CAD from other companies.
If exported as a DICOM-RT Structure Set, it can be used to evaluate lung function in iVAS, our OEM product, or displayed in the free version of Growth RTV Edu.
*This software is not certified as a medical device. Do not use it for medical treatment such as diagnosis or therapy.
For organs not supported by the basic functionality, we can customize the system to meet the needs of the medical or research facility of your choice, with additional training for region extraction using image data from the facility.
Please consult with us regarding the form of implementation and fees for additional study.

Our proprietary “Deep Seal ®” technology (patent pending) for identification and protection of deep learning models makes it possible to characterize deep learning models by having them store special inputs and outputs during training.
This technology can be used to characterize not only the basic models installed in Growth RTV, but also models that have been customized and learned at the facility where they are installed, thereby preventing unauthorized use and diversion of learned models through code leakage and reverse engineering.
In addition, multiple patterns can be stored, not only a single pattern, if desired, leading to improved deterrence.
Graphics Memory 2.0 GB or more (4.0 GB or more recommended)
| ITEM | REQUIREMENTS |
|---|---|
| OS | Windows 10/11 (64bit) |
| CPU/Memory | Intel/Ryzen 4-Core CPU or higher / 8GB or higher |
| Graphics | GPUs with NVIDIA GeForce GTX400 series chipsets or later that support CUDA technology |
| Supported format | DICOM/DICOM-RT |
| Compatible CT and MRI images | Uncompressed/Jpeg Lossless format/Jpeg 2000 format |
| Supported Transfer Protocol | DICOM Query/Retrieve |
1) This product is not certified as a medical device. Do not use for medical treatment such as diagnosis or therapy.
 Return to Top ▲
Return to Top ▲